MerlinViewer
About this program
When writing mapping software I analyse small test pedigrees with the program I
am working on and other mapping software like Merlin. However, when using the
Merlin software and high density SNP data, the output
data consists of a very large results table which due to its size may be difficult to
process by eye. To over come this problem MerlinViewer was developed to visualise the
data as a scatter graph of the LOD, HLOD or Alpha score against chromosomal position.
The MerlinViewer program first reads the appropriate map file and then the Merlin
output text file. After pressing the 'View' button the data is read and displayed
as a scatter graph with the x-axis representing the SNPs position and the y-axis
representing the SNPs linkage score.
Data from a region can be exported as either a tab delimited text file or a html
web page. To export data, place the cursor at one side of the region and with the mouse
button pressed move the cursor to the other side of the region. Then press the
Save button and select the correct file extension and enter
the name of the file to save the data too.
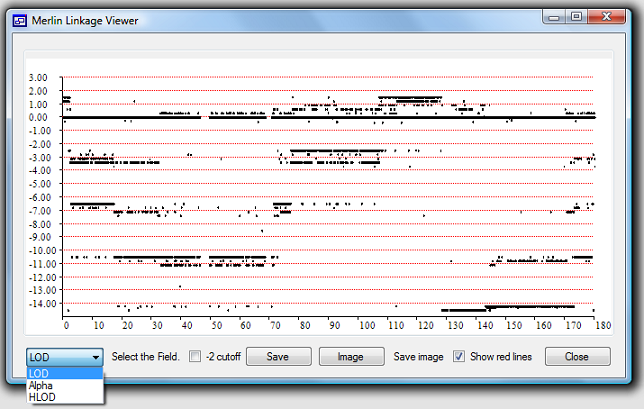
Figure 1
Each *.map
and merlin data file must contain the data for a single chromosome and be the standard format
as exported by the program.
Citation
This was not published in its own right, but was mentioned in this paper:
Carr IM, Johnson CA, Markham AF, Tomes C, Bonthron DT, Sheridan EG.
DominantMapper: Rule-based analysis of SNP data for rapid mapping of dominant diseases in related nuclear families.
Hum Mutat. Accepted 2011 Aug 11
 MerlinViewer and a set of map and
results files can be downloaded from my works web page here
MerlinViewer and a set of map and
results files can be downloaded from my works web page here
Note: this program requires the .NET framework version 2.0 to be installed
(available through Windows Update or
here).
- Merlin details
- Merlin was developed by GR Abecasis at the Center for Statistical Genetics,
University of Michigan and has it own web page
here
- Merlin citation
- Abecasis GR, Cherny SS, Cookson WO and Cardon LR (2002).
Merlin-rapid analysis of dense genetic maps using sparse gene flow trees.
Nature Genetics 30:97-101.
|