Jukes-Cantor genetic distances
Purpose of this page
This page calculates the genetic distance and its variance between two sequences using
the Jukes-Cantor model. Rather than create the alignment, it is assumed that you have done
this and already know the number of changes and the length of the sequences.
The genetic distance between two sequences is the sum of the substitutions that have
accumulated between them since they diverged from their common ancestor. While it is possible
to count the differences between the two sequences, this would under estimate the true divergence
since some positions may have undergone multiple changes. To overcome this problem Charles
Cantor and Thomas Jukes developed a probabilistic model and derived the following formula to
calculate genetic distances:
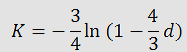
where K is the genetic distance and d is the number of observed substitutions divided by the sequence length. They also found the variance of K could be found by:
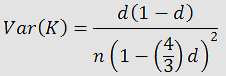
 A Windows program that duplicates this page can be downloaded here.
A Windows program that duplicates this page can be downloaded here.
|