IBDelphi populations was developed as an extension to IBDelphi Duo,
so that it is possible to automatically perform non-redundant pair wise analysis of genotype data from a large number of individuals,
from one or two distinct populations.
Genotyping should be performed using very high density SNP microarrays such as Affymetrix
SNP5 or SNP6 chips. These data files must be annotated with chromosome and positional data, which
can conveniently be done using SNP6Annotator.
Computer requirements
This program is designed to run on Windows XP SP3 ,Vista SP1 and Windows 7 systems that have the .NET 2.0 framework installed,
which is freely available from
Microsoft .
Data analysis
Inter-population analysis
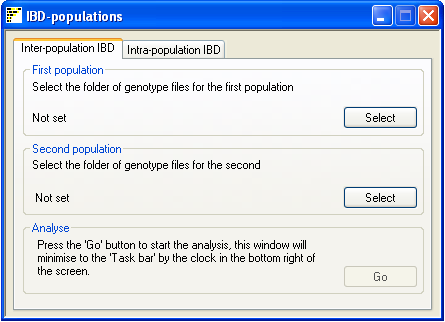
Figure 1
When analysing data from two populations, place the genotype files in two folders so that each folder contains data for
only one population. Next select each folder in turn using the Select
buttons in the First population and Second population panels (figure 1).
After both folders have been selected the Go button in the Analyse panel
will be activated. Pressing this button will prompt the user for the name of the results file and start the analysis.
IBDeplhi populations will compare each genotype data file in the first folder to each data file in the second folder. Since
the analysis may take some time IBDelphi populations automatically minimises to an icon in the taskbar
(figure 2a). To restore the IBDelphi populations window right click its icon in the taskbar and select
Show (figure 2b), because IBDelhi populations is analysing the data it may be slow
to respond. Selecting the Quit option will save the present analysis results and then close
IBDElphi populations.
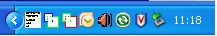 |
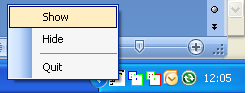 |
a |
b |
Figure 2
Intra-population analysis
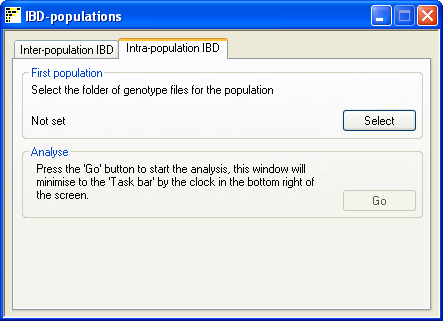
Figure 3
To analyse data form a single population select the Intra-population IBD tab option (figure 3) and enter the
folder containing the genotype data. The Go button in the Analyse panel will be
activated, pressing it will prompt the user to enter the name of the results file and then start the analysis. IBDelphi
population will then compare each pair of data files in the folder to find the total length of IBD between the two individuals and
save the result to the results file. Since the analysis may take some time then IBDelphi populations
automatically minimises to an icon in the taskbar (figure 2). To restore the IBDelphi populations window
right click its icon in the taskbar and select Show, because IBDelhi populations
is analysing the data it may be slow to respond.
Viewing the results of the analysis
Since it is expected that the results from the analysis will be analysed in a number of different ways, the export file structure has been kept simple
with both analysis options creating tab delimited text files which can be opened in a spread sheet program such as Excel. In each, the first
column lists the total length of IBD (in Mb's) between the two files listed in the second and third columns. IBDelphi population reads the
file in column 2 once and then compares it to all the other files listed in column 3, consequently the data is grouped around the file in second
column. Tables 1 and 2 show the first six rows of data from a inter-population and intra-population analysis respectively.
| Total length of IBD (Mb) | File from first population | File from second population |
|---|
| 10,294,540 | NA06984 CEU.birdseed-v2_anot.xls | NA18939 JPT.birdseed-v2_anot.xls |
| 5,369,343 | NA06984 CEU.birdseed-v2_anot.xls | NA18946 JPT.birdseed-v2_anot.xls |
| 4,299,804 | NA06984 CEU.birdseed-v2_anot.xls | NA18954 JPT.birdseed-v2_anot.xls |
| 3,715,222 | NA06984 CEU.birdseed-v2_anot.xls | NA18955 JPT.birdseed-v2_anot.xls |
| 4,353,238 | NA06984 CEU.birdseed-v2_anot.xls | NA18957 JPT.birdseed-v2_anot.xls |
| 0 | NA06984 CEU.birdseed-v2_anot.xls | NA18962 JPT.birdseed-v2_anot.xls |
Table 1
| Total length of IBD (Mb) | File A | File B |
|---|
| 3,520,845 | NA19625 ASW.birdseed-v2_anot.xls | NA19700 ASW.birdseed-v2_anot.xls |
| 0 | NA19625 ASW.birdseed-v2_anot.xls | NA19701 ASW.birdseed-v2_anot.xls |
| 12,600,070 | NA19625 ASW.birdseed-v2_anot.xls | NA19703 ASW.birdseed-v2_anot.xls |
| 5,852,790 | NA19625 ASW.birdseed-v2_anot.xls | NA19704 ASW.birdseed-v2_anot.xls |
| 0 | NA19625 ASW.birdseed-v2_anot.xls | NA19711 ASW.birdseed-v2_anot.xls |
Table 2